Overview#
This package is a complete tool for the analysis of cell morphology in time-lapse microscopy acquisitions. It’s main purpose is to enable the study of proteins fluxes, via by GFP tagging, by subdividing the observed cell into local “windows” that are tracked over time. The package consist of two parts: a first part handling cell segmentation, windowing and tracking over time; and a second part allowing to analyze the content of windows over time and to correlate it with cell dynamics. To run the software we provide Jupyter-based interfaces, but the package can also be used separately from the UI.
This package is the result of a collaboration between the Microscopy Imaging Center, the Olivier Pertz lab and Science IT Support at Bern University and has been developed by Cédric Vonesch (ScITS, Pertz lab) and Guillaume Witz (MIC, ScITS) with the collaboration of Jakobus van Unen (Pertz lab).
Installation#
You can find installation instruction here. We rely on conda for installation, so if you don’t have some form of conda already installed, we recommend installing miniconda by following these instructions. Installation works in the same way on a laptop and on a cluster.
Most of the computationally heavy aspect of the analysis are parallelized by using Dask, and functions in exactly the same way on a laptop or on a cluster, and as described in detail in later sections, one has just to specify which option is used.
What data can be processed#
A typical dataset consists in a multi-channel time-lapse acquisition. Cells to be analyzed should ideally not be touching and not drift out of the field of view.
At the moment four types of datasets are supported:
Each channel is stored in a separate folder which contains a series of tiff files, one per time point.
All channels are individually stored as a multi-page tiff stack and located in a single folder
The data are stored in Nikon’s ND2 format (this is very experimental).
All channels are individually stored as h5 files and located in a single folder
In the package the datasets are handled by the dataset module.
Typical analysis pipeline#
The analysis proceeds in the following way:
The images are segmented, i.e. cells are detected and a binary mask is created for each image. There are multiple options for segmentation, and in principle you can add additional custom function in the code.
The cell is tracked across frames. This allows to handle images with more than one cell.
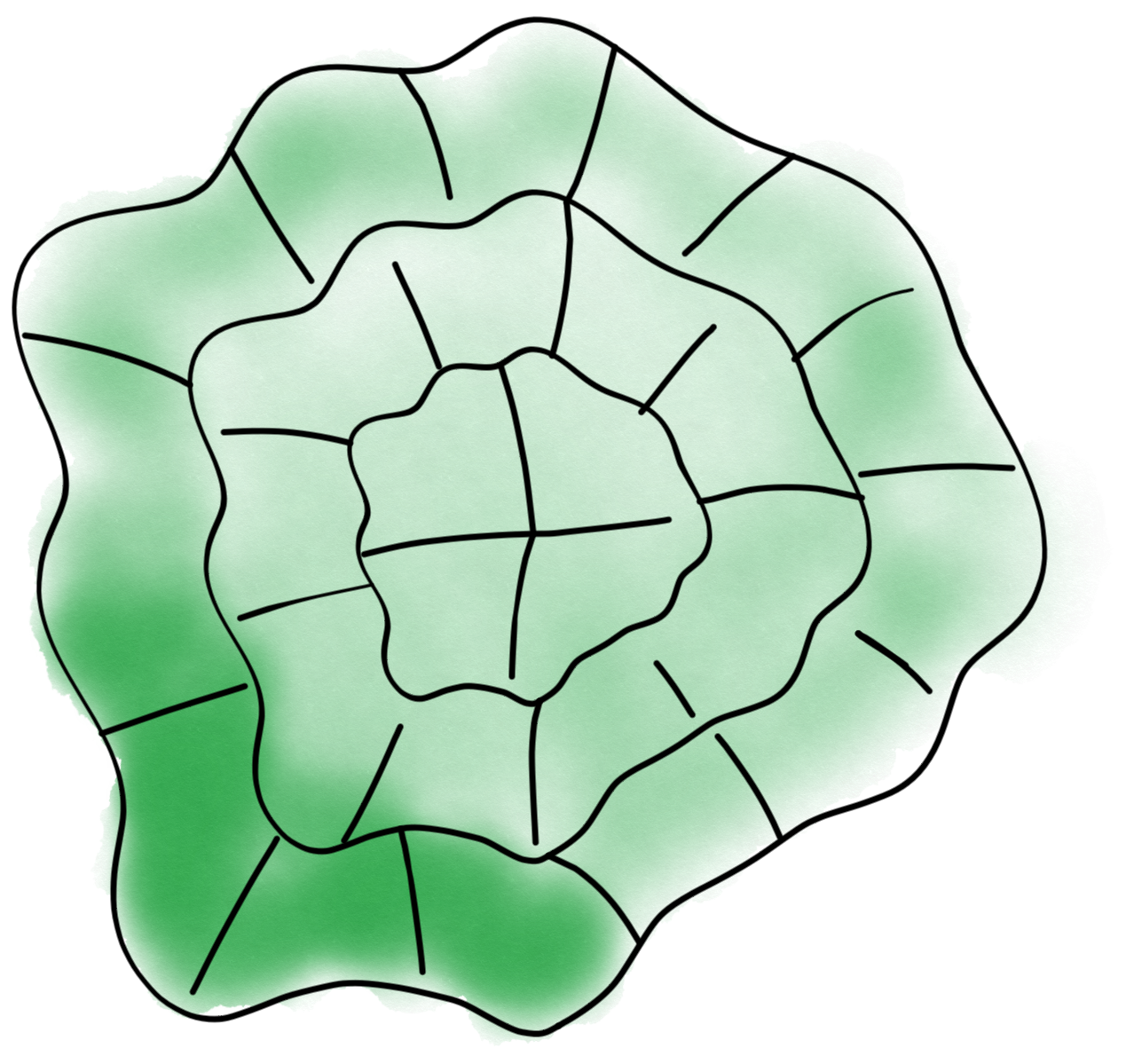
A continuous spline is fitted to the cell in all frames and the the cell area is split into small “windows”. In this step, great care is taken to “align” successive contours so as to best estimate deformation and ensure that the position of windows is not drifting.
Windows and contours can be used to extract information about fluorescence and contour dynamics.
To execute the pipeline, you can either use the napari plugin, the interfaced provided in the notebook or use the package as an API. You can find more information one the whole analysis process in the following sections:
Running an analysis with the notebook UI
Running an analysis without UI