Projects
Coupling of cell division and DNA replication in E.coli
My current project funded by an SNF Ambizione fellowship is centered around the bacterial cell cycle. In order to achieve robust growth in various conditions, bacteria need to coordinate their division and DNA replication cycles. A powerful way to get insight into that coordination and to infer conceptual regulatory strategies is to observe specific events related to each cycle in complete cell cycles of living bacteria. I achieve this by growing E.coli in microfluidics devices and using phase contrast and fluorescence microscopy to track these events. The movie below shows an example of the type of microscopy data that I am acquiring. Cells can be seen in dark blue, growing in horizontal microfluidic channels. The origin of replication is tagged and appears as red dots. The division machinery is made visible through the tagging of the FtsZ protein in green. I designed software to detect and track those elements automatically in each cell. For example the tracking of the red dots (origin of replication) and the detection of the moment where those split into two, allows to identify a replication initiation event.
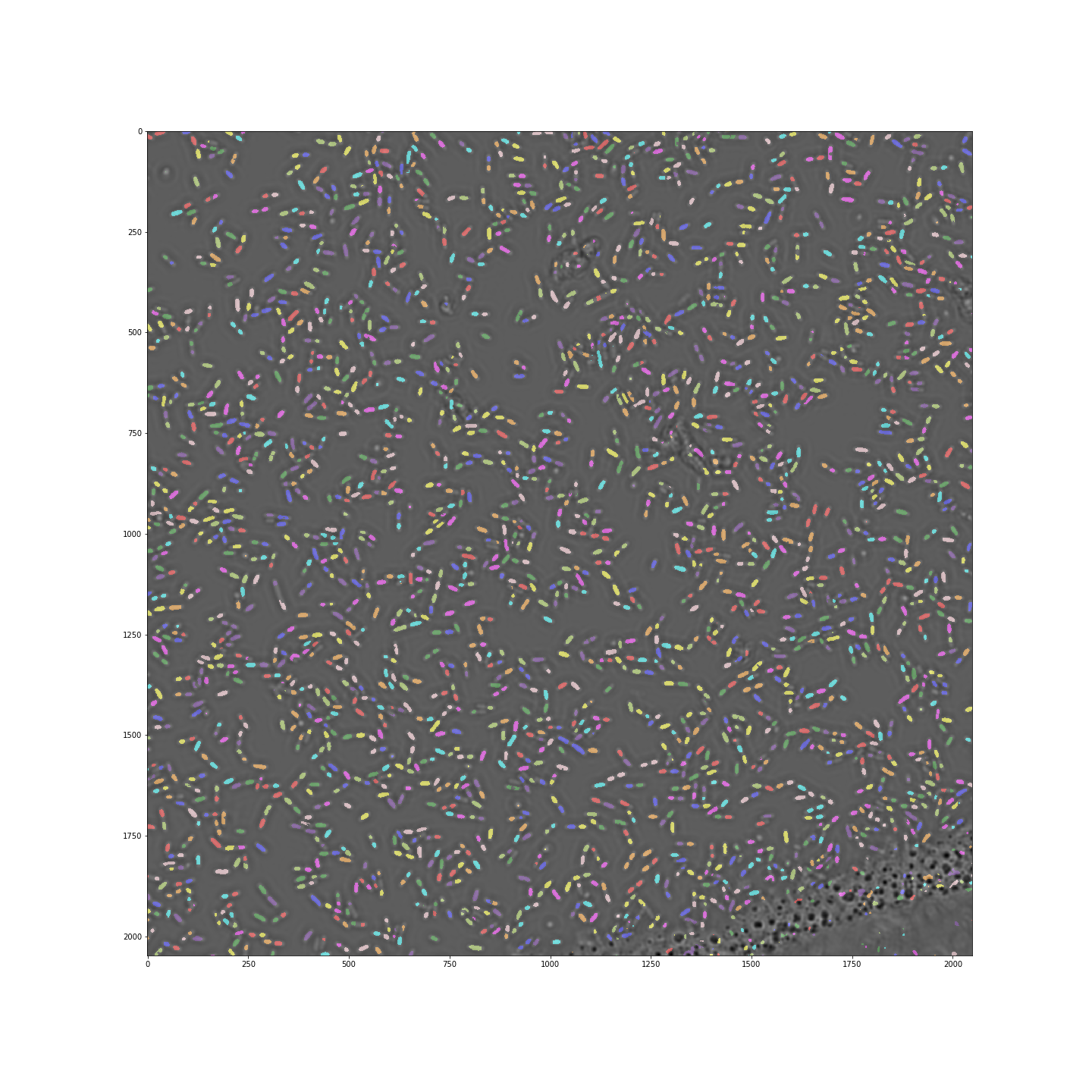
Deep Learning for image segmentation
Deep learning methods have received a lot of attention in the past years. While in many cases simpler methods works as well, image analysis is the one application where deep learning shines. Using tools such as Keras and Tensorflow which simplify access to deep learning methods, I am exploring the feasibility to use those to perform routine image segmentation in various microscopy based projects. The image below shows how a u-net convolutional network achieves excellent segmentation of dense E.coli cells imaged over a large area in a 96-well plate (see code here). The video shows that deep learning can be applied successfully in very different situations. On the left cells growing in a microfluidic channel are segmented, while on the right fluorescent spots within the cells (tagged origin of replication) are segmented.
 ..
..
Design and usage of new microfluidics design
Performing biology experiments in microfluidic devices remains a somewhat challenging task because of multiple technical barriers. In collaboration with Thomas Julou (UniBas), we put a constant effort in trying to improve our methods to make such experiments more accessible. One such effort is focusing on improving the design of the mother machine, a device in which cells grow in lineages in narrow channels closed at one end. One systematic problem is the loading of cells in such devices which mostly relies on diffusion. We developed a new version of that device by adding a small opening at the closed end of the growth channels and exploiting a pressure system to actively drive cells into them. The details of the device are not yet made publicly available. However, to develop that device I also wrote a small python library to create multi-layer microfluidics designs. It is still experimental but available here. I also made available a minimal example of how to write a Jupyter file to generate a new design. If you want to use the module but are confused, don’t hesitate to get in touch.
Hardware synchronization
In order to be able to acquire long time-lapses of growing cells with regular fluorescence microscopy acquisitions, one has to minimize has much as possible photodamage. In particular, one can optimize a microscopy setup in such a way that the sample is only illuminated when the camera is actually recording an image. This is typically not achieved with conventional setups where a computer serves as central command and issues order to the different microscopy parts, introducing relatively long delays e.g. between turning illumination ON and starting camera recording. A way to go around that is to use the camera as central command, in a so-called hardware synchronization mode. In collaboration with Thomas Julou (UniBas) we wrote a detailed description of how to achive such hardware triggering based on open source resources such as MicroManager and Arduino.
Past projects
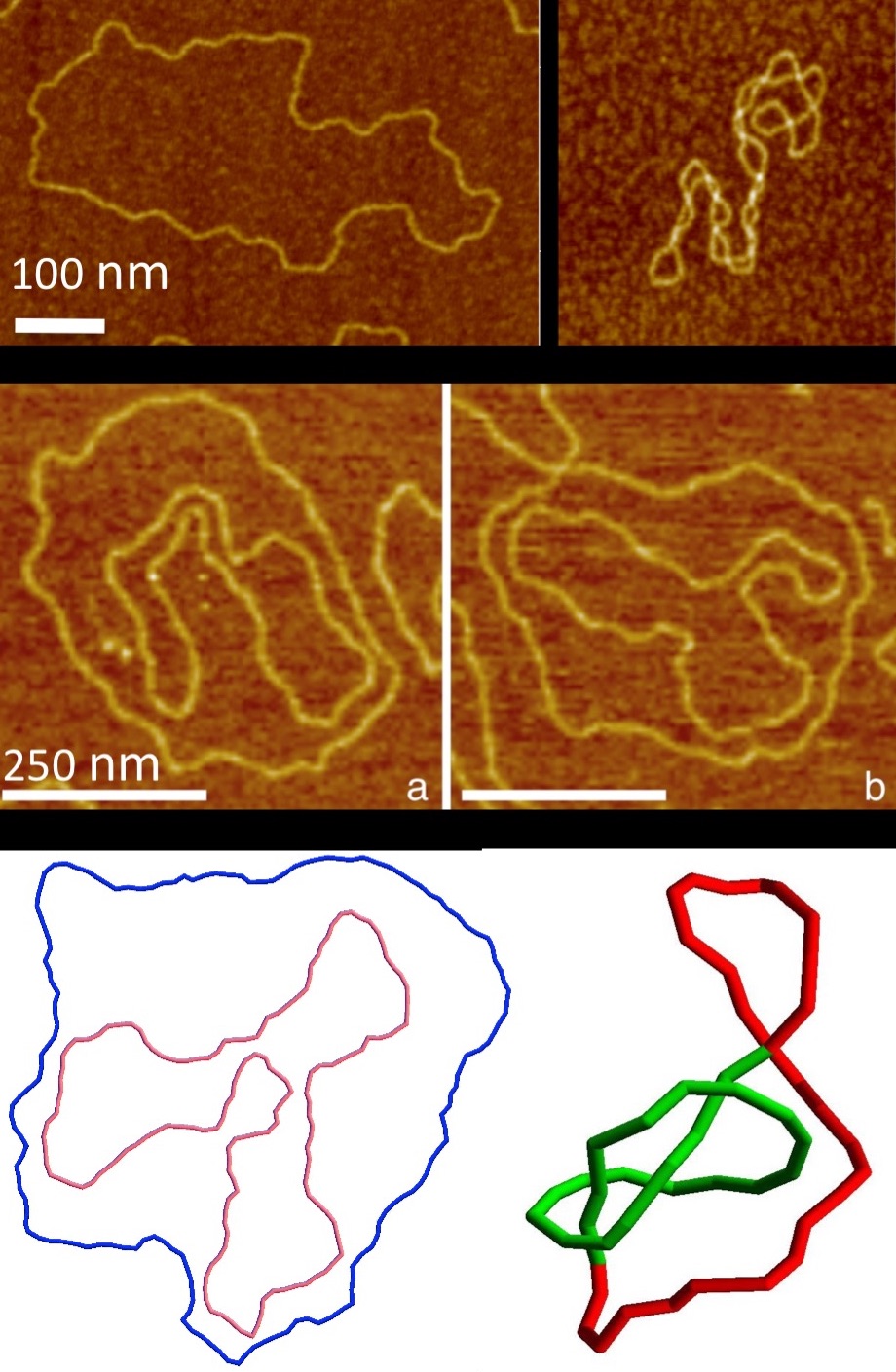
DNA topology
My Ph.D. thesis was centered around DNA topology following two axis: 1) a polymer physics perspective where I imaged DNA by AFM and used it as a model to study topological effects in polymer physics, and 2) a biophysical perspective where I studied mainly through numerical simulations the the biological consequences of topological effects like supercoiling and knotting.
 ..
..
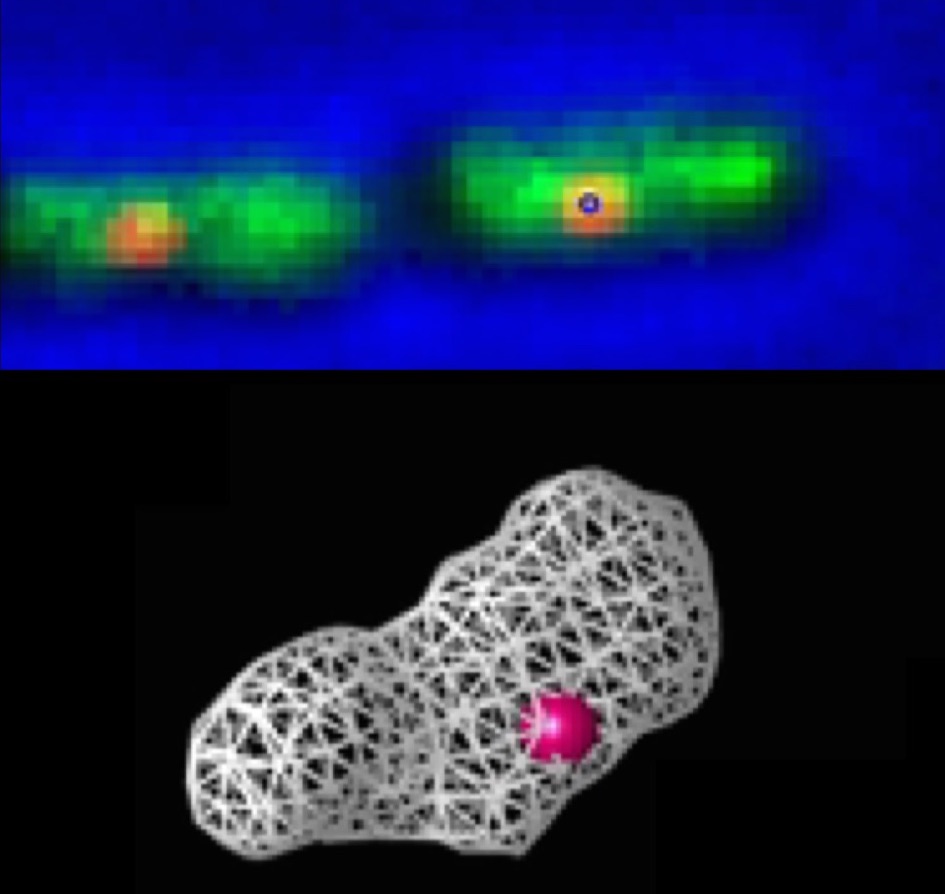
E.coli nucleoid imaging
During my postdoc at Harvard University, I used fluorescence microscopy to study the structure and dynamics of the E.coli nucleoid. By acquiring images of the nucleoid tagged with a GFP-fusion of a DNA binding protein (HupA) at several focal planes, one can reconstruct its 3D shape. Additionally, the origin of replication was also tagged, allowing to study the relation between the dynamics of a single locus and the global chromosome dynamics. See here for more details.
 ..
..