What is napari-morphodynamics?#
napari-morphodynamics is a plugin that allows to study the dynamics of cell morphology and fluorescence intensity fluctuations over time in 2D single-cell time lapse images. Such analysis comes with different challenges as it relies on 1) on an accurate cell segmentation over time, 2) a specific partition of the cell in different regions of interest (windows) that should be conserved over time and in which intensity can be analysed, 3) the analysis of cell shape, window intensity and the correlation between these variables.
napari-morphodynamics gives a user-friendly way to perform all these steps in one place. We briefly review them here, and indicate where to find more information.
Walk trough#
Data sources#
The plugin allows to import various standard formats of data, including series of tifs, multipage tifs, and zarr. So that the plugin can function correctly, these data should come in a specific format. For tif formats you can refer to this page. For numpy or zarr arrays, the expected dimensions are CTYX.
As it can be difficult to provide the correct format and some manul handling might be necessary in napari, we also offer the possibility to use data from the napari layers as input. In that case, the selected layers are saved as zarr files and handled normall after that. Please refer to the specific example.
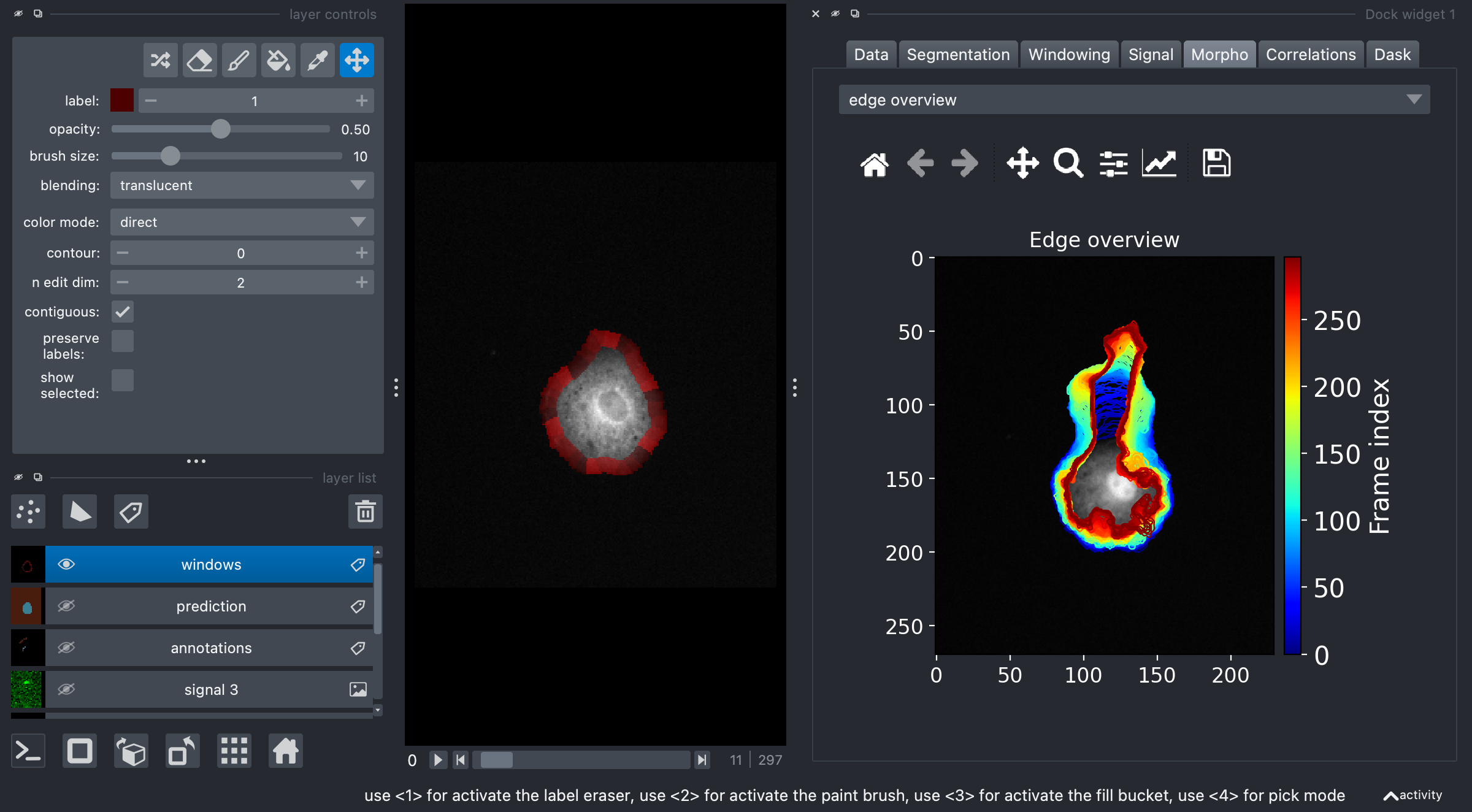
Segmentation#
Segmentation can also be performed in multiple ways. By default the plugin will off the napari-convpaint solution, based on another plugin. The idea is here to provide example annotations of background and foreground in order to train a segmentation model. You can find an example here.
Alternatively, we give access to cellpose and its cyto2 model (more models may be added later). In order to use cellpose, you need to install cellpose in the same environment or use directly pip install "napari-morphodynamics[cellpose]" for plugin installation. Finally,
Finally, via the precomputed option, one can either re-use a segmentation performed with the plugin or use a layer created in other ways, e.g. manually
Windowing#
Windowing is the operation of splitting a segmentation into smaller parts whose integrated intensity can then be tracked. The only options to specify here are the desired window size and smoothness of the segmentation contour. You can find further detailed information on window creation in the MorphoDynamics package documentation.
Post-processing#
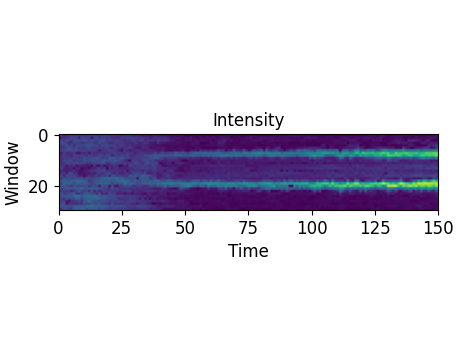
We offer three types of data representation. First one can display the intensity per window over time:
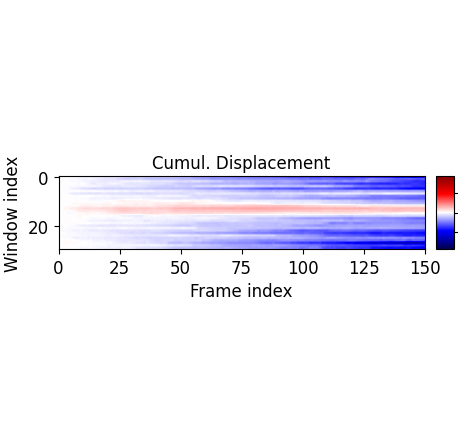
Second, one can display various displacement variables such as the cumulative displacement:
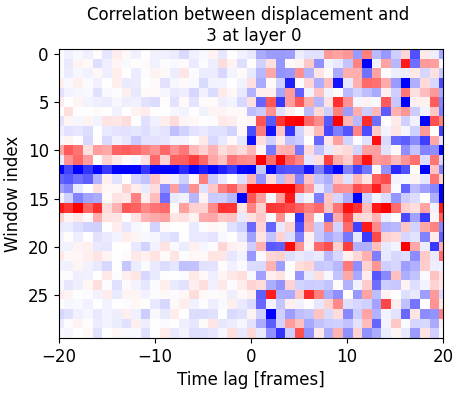
Finally we can look at the time correlation between signals or between cell edge movement and signals: