Advanced processing¶
In the previous notebook, we have seen a very simple analysis pipeline. The great advantage of using Python is however that you get access to cutting edge method relatively easily. As you can also run regular commands from the notebook, you can also mix different softwares directly in your pipeline.
We illustrate these two points here with Cellpose, a generalistic deep learning based cell segmentation approach and Ilasitk, the popular software for ML based segmentation.
Dataset¶
First we re-load our dataset:
import requests
import zipfile
from pathlib import Path
if not Path('BBBC013_v1_images_bmp').is_dir():
url = 'https://data.broadinstitute.org/bbbc/BBBC013/BBBC013_v1_images_bmp.zip'
myfile = requests.get(url, allow_redirects=True)
open('BBBC013_v1_images_bmp.zip', 'wb').write(myfile.content)
with zipfile.ZipFile('BBBC013_v1_images_bmp.zip', 'r') as zip_ref:
zip_ref.extractall('.')
data_path = Path('BBBC013_v1_images_bmp')
file_list2 = list(data_path.glob('Channel2*.BMP'))
Deep Learning made easy: example with cellpose¶
Cellpose is a deep learning algorithm that has been trained on a massive trove of data to identify nuclei and cells. The variety in the training set makes it extremely versatile so that retrainig is not necessary in most standard cases.
Installing cellpose¶
Unlike Numpy, scikit-image etc. cellpose is not available by default on Colab, so you have to install it via pip:
%%bash
pip install cellpose
pip install git+https://github.com/guiwitz/microfilm.git@master#egg=microfilm
pip install imagecodecs
Requirement already satisfied: cellpose in /usr/local/lib/python3.7/dist-packages (0.6.1)
Requirement already satisfied: tifffile in /usr/local/lib/python3.7/dist-packages (from cellpose) (2021.4.8)
Requirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from cellpose) (1.19.5)
Requirement already satisfied: opencv-python-headless in /usr/local/lib/python3.7/dist-packages (from cellpose) (4.5.1.48)
Requirement already satisfied: torch>=1.6 in /usr/local/lib/python3.7/dist-packages (from cellpose) (1.8.1+cu101)
Requirement already satisfied: natsort in /usr/local/lib/python3.7/dist-packages (from cellpose) (5.5.0)
Requirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from cellpose) (4.41.1)
Requirement already satisfied: numba in /usr/local/lib/python3.7/dist-packages (from cellpose) (0.51.2)
Requirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from cellpose) (1.4.1)
Requirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from torch>=1.6->cellpose) (3.7.4.3)
Requirement already satisfied: llvmlite<0.35,>=0.34.0.dev0 in /usr/local/lib/python3.7/dist-packages (from numba->cellpose) (0.34.0)
Requirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from numba->cellpose) (54.2.0)
Collecting microfilm
Cloning https://github.com/guiwitz/microfilm.git (to revision master) to /tmp/pip-install-jxz0mzdt/microfilm
Installing build dependencies: started
Installing build dependencies: finished with status 'done'
Getting requirements to build wheel: started
Getting requirements to build wheel: finished with status 'done'
Preparing wheel metadata: started
Preparing wheel metadata: finished with status 'done'
Requirement already satisfied: matplotlib in /usr/local/lib/python3.7/dist-packages (from microfilm) (3.2.2)
Requirement already satisfied: h5py in /usr/local/lib/python3.7/dist-packages (from microfilm) (2.10.0)
Collecting aicsimageio
Downloading https://files.pythonhosted.org/packages/d2/83/539227400db61fd6ebecbb5305dcde24783f9422e50962e8a202eebea056/aicsimageio-3.3.4-py2.py3-none-any.whl (60kB)
Requirement already satisfied: tifffile in /usr/local/lib/python3.7/dist-packages (from microfilm) (2021.4.8)
Requirement already satisfied: scikit-image in /usr/local/lib/python3.7/dist-packages (from microfilm) (0.16.2)
Collecting nd2reader@ git+https://github.com/guiwitz/nd2reader.git@master#egg=nd2reader
Cloning https://github.com/guiwitz/nd2reader.git (to revision master) to /tmp/pip-install-jxz0mzdt/nd2reader
Requirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from microfilm) (1.19.5)
Requirement already satisfied: natsort in /usr/local/lib/python3.7/dist-packages (from microfilm) (5.5.0)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib->microfilm) (0.10.0)
Requirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->microfilm) (2.8.1)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->microfilm) (1.3.1)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->microfilm) (2.4.7)
Requirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from h5py->microfilm) (1.15.0)
Collecting lxml>=4.4.2
Downloading https://files.pythonhosted.org/packages/cf/4d/6537313bf58fe22b508f08cf3eb86b29b6f9edf68e00454224539421073b/lxml-4.6.3-cp37-cp37m-manylinux1_x86_64.whl (5.5MB)
Requirement already satisfied: toolz>=0.10.0 in /usr/local/lib/python3.7/dist-packages (from aicsimageio->microfilm) (0.11.1)
Requirement already satisfied: imageio>=2.3.0 in /usr/local/lib/python3.7/dist-packages (from aicsimageio->microfilm) (2.4.1)
Collecting aicspylibczi<3.0,>=2.7.5
Downloading https://files.pythonhosted.org/packages/06/aa/2e238f68434731561c1b84701e885c537e237dccf13f51818659704b8c93/aicspylibczi-2.8.0-cp37-cp37m-manylinux2010_x86_64.whl (621kB)
Collecting imagecodecs>=2020.5.30
Downloading https://files.pythonhosted.org/packages/d3/b7/b376a1e8d3c7f7f83012e722a9c11606137540248f89766bb28a8dc879d4/imagecodecs-2021.3.31-cp37-cp37m-manylinux2014_x86_64.whl (24.7MB)
Collecting distributed>=2.9.3
Downloading https://files.pythonhosted.org/packages/0c/b0/3454dc44239c526f9c9e4cf04f62823776b71f927db74302986d56e7a9a1/distributed-2021.4.0-py3-none-any.whl (684kB)
Collecting readlif>=0.2.1
Downloading https://files.pythonhosted.org/packages/4d/b9/1e39639a06659dadde0fff60b6b496600fbfd2b519174f0930f155bcb0b4/readlif-0.5.1-py3-none-any.whl
Requirement already satisfied: dask>=2.9.0 in /usr/local/lib/python3.7/dist-packages (from aicsimageio->microfilm) (2.12.0)
Requirement already satisfied: pillow>=4.3.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image->microfilm) (7.1.2)
Requirement already satisfied: networkx>=2.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image->microfilm) (2.5.1)
Requirement already satisfied: PyWavelets>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image->microfilm) (1.1.1)
Requirement already satisfied: scipy>=0.19.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image->microfilm) (1.4.1)
Collecting xmltodict>=0.9.2
Downloading https://files.pythonhosted.org/packages/28/fd/30d5c1d3ac29ce229f6bdc40bbc20b28f716e8b363140c26eff19122d8a5/xmltodict-0.12.0-py2.py3-none-any.whl
Collecting pims>=0.3.0
Downloading https://files.pythonhosted.org/packages/d5/47/82e0ac31e01a271e5a06362fbf03769e9081956f6772f91d98b32899d743/PIMS-0.5.tar.gz (85kB)
Requirement already satisfied: psutil>=5.0 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (5.4.8)
Requirement already satisfied: sortedcontainers!=2.0.0,!=2.0.1 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (2.3.0)
Requirement already satisfied: pyyaml in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (3.13)
Requirement already satisfied: tornado>=5; python_version < "3.8" in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (5.1.1)
Requirement already satisfied: click>=6.6 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (7.1.2)
Requirement already satisfied: zict>=0.1.3 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (2.0.0)
Requirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (54.2.0)
Collecting cloudpickle>=1.5.0
Downloading https://files.pythonhosted.org/packages/e7/e3/898487e5dbeb612054cf2e0c188463acb358167fef749c53c8bb8918cea1/cloudpickle-1.6.0-py3-none-any.whl
Requirement already satisfied: tblib>=1.6.0 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (1.7.0)
Requirement already satisfied: msgpack>=0.6.0 in /usr/local/lib/python3.7/dist-packages (from distributed>=2.9.3->aicsimageio->microfilm) (1.0.2)
Requirement already satisfied: decorator<5,>=4.3 in /usr/local/lib/python3.7/dist-packages (from networkx>=2.0->scikit-image->microfilm) (4.4.2)
Collecting slicerator>=0.9.8
Downloading https://files.pythonhosted.org/packages/75/ae/fe46f5371105508a209fe6162e7e7b11db531a79d2eabcd24566b8b1f534/slicerator-1.0.0-py3-none-any.whl
Requirement already satisfied: heapdict in /usr/local/lib/python3.7/dist-packages (from zict>=0.1.3->distributed>=2.9.3->aicsimageio->microfilm) (1.0.1)
Building wheels for collected packages: microfilm
Building wheel for microfilm (PEP 517): started
Building wheel for microfilm (PEP 517): finished with status 'done'
Created wheel for microfilm: filename=microfilm-0.0.2.dev11+g06a762d-cp37-none-any.whl size=9802 sha256=7d1a1a757d8deb9b46361c7a2b55cb9259cec6c18870f0267c8b4be20fd988ed
Stored in directory: /tmp/pip-ephem-wheel-cache-zlgct8oy/wheels/d0/c4/b6/02340665da49a886bb5a33201402799512e530c6fac55fd0fe
Successfully built microfilm
Building wheels for collected packages: nd2reader, pims
Building wheel for nd2reader (setup.py): started
Building wheel for nd2reader (setup.py): finished with status 'done'
Created wheel for nd2reader: filename=nd2reader-3.2.3.gw-py2.py3-none-any.whl size=35972 sha256=69afb49a556d307e0fcb5cca5f859454cae19251ec17ceb0af1409d75fa5770c
Stored in directory: /tmp/pip-ephem-wheel-cache-zlgct8oy/wheels/da/50/c3/8a18ff8641c808ee4af091f0264d2b68e37745472308ffafb9
Building wheel for pims (setup.py): started
Building wheel for pims (setup.py): finished with status 'done'
Created wheel for pims: filename=PIMS-0.5-cp37-none-any.whl size=84328 sha256=8b89c5d751663effda88fd6541043c8c1e58bbc8bb2e4e05e643b7a8353dc54e
Stored in directory: /root/.cache/pip/wheels/0e/0a/14/4c33a4cc1b9158e57329a38e8e3e03901ed24060eb322d5462
Successfully built nd2reader pims
Installing collected packages: lxml, aicspylibczi, imagecodecs, cloudpickle, distributed, readlif, aicsimageio, xmltodict, slicerator, pims, nd2reader, microfilm
Found existing installation: lxml 4.2.6
Uninstalling lxml-4.2.6:
Successfully uninstalled lxml-4.2.6
Found existing installation: cloudpickle 1.3.0
Uninstalling cloudpickle-1.3.0:
Successfully uninstalled cloudpickle-1.3.0
Found existing installation: distributed 1.25.3
Uninstalling distributed-1.25.3:
Successfully uninstalled distributed-1.25.3
Successfully installed aicsimageio-3.3.4 aicspylibczi-2.8.0 cloudpickle-1.6.0 distributed-2021.4.0 imagecodecs-2021.3.31 lxml-4.6.3 microfilm-0.0.2.dev11+g06a762d nd2reader-3.2.3.gw pims-0.5 readlif-0.5.1 slicerator-1.0.0 xmltodict-0.12.0
Running command git clone -q https://github.com/guiwitz/microfilm.git /tmp/pip-install-jxz0mzdt/microfilm
Running command git clone -q https://github.com/guiwitz/nd2reader.git /tmp/pip-install-jxz0mzdt/nd2reader
Running command git submodule update --init --recursive -q
ERROR: distributed 2021.4.0 has requirement dask>=2021.03.0, but you'll have dask 2.12.0 which is incompatible.
Checking GPU¶
As cellpose is deep-learning based, it can exploit the powere of GPUs to run. On Colab you can choose to use a GPU by changing the runtime type in the Runtime menu. Check if the GPU is used:
!nvcc --version
!nvidia-smi
nvcc: NVIDIA (R) Cuda compiler driver
Copyright (c) 2005-2020 NVIDIA Corporation
Built on Wed_Jul_22_19:09:09_PDT_2020
Cuda compilation tools, release 11.0, V11.0.221
Build cuda_11.0_bu.TC445_37.28845127_0
Sat Apr 17 18:48:07 2021
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 460.67 Driver Version: 460.32.03 CUDA Version: 11.2 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 Tesla T4 Off | 00000000:00:04.0 Off | 0 |
| N/A 43C P8 9W / 70W | 0MiB / 15109MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| No running processes found |
+-----------------------------------------------------------------------------+
Load and use a model¶
Now we can load a cellpose model and use it to segment the nuclei of our image:
from cellpose import models
model = models.Cellpose(gpu=True, model_type='nuclei')
Downloading: "https://www.cellpose.org/models/cyto_0" to /root/.cellpose/models/cyto_0
100%|██████████| 25.2M/25.2M [00:00<00:00, 79.4MB/s]
Downloading: "https://www.cellpose.org/models/cyto_1" to /root/.cellpose/models/cyto_1
100%|██████████| 25.2M/25.2M [00:00<00:00, 89.2MB/s]
Downloading: "https://www.cellpose.org/models/cyto_2" to /root/.cellpose/models/cyto_2
100%|██████████| 25.2M/25.2M [00:00<00:00, 96.0MB/s]
Downloading: "https://www.cellpose.org/models/cyto_3" to /root/.cellpose/models/cyto_3
100%|██████████| 25.2M/25.2M [00:00<00:00, 96.2MB/s]
Downloading: "https://www.cellpose.org/models/size_cyto_0.npy" to /root/.cellpose/models/size_cyto_0.npy
100%|██████████| 3.55k/3.55k [00:00<00:00, 8.82MB/s]
Downloading: "https://www.cellpose.org/models/cytotorch_0" to /root/.cellpose/models/cytotorch_0
100%|██████████| 25.3M/25.3M [00:00<00:00, 95.1MB/s]
Downloading: "https://www.cellpose.org/models/cytotorch_1" to /root/.cellpose/models/cytotorch_1
100%|██████████| 25.3M/25.3M [00:00<00:00, 99.1MB/s]
Downloading: "https://www.cellpose.org/models/cytotorch_2" to /root/.cellpose/models/cytotorch_2
100%|██████████| 25.3M/25.3M [00:00<00:00, 104MB/s]
Downloading: "https://www.cellpose.org/models/cytotorch_3" to /root/.cellpose/models/cytotorch_3
100%|██████████| 25.3M/25.3M [00:00<00:00, 95.6MB/s]
Downloading: "https://www.cellpose.org/models/size_cytotorch_0.npy" to /root/.cellpose/models/size_cytotorch_0.npy
100%|██████████| 5.23k/5.23k [00:00<00:00, 4.36MB/s]
Downloading: "https://www.cellpose.org/models/nuclei_0" to /root/.cellpose/models/nuclei_0
100%|██████████| 25.2M/25.2M [00:00<00:00, 83.8MB/s]
Downloading: "https://www.cellpose.org/models/nuclei_1" to /root/.cellpose/models/nuclei_1
100%|██████████| 25.2M/25.2M [00:00<00:00, 60.0MB/s]
Downloading: "https://www.cellpose.org/models/nuclei_2" to /root/.cellpose/models/nuclei_2
100%|██████████| 25.2M/25.2M [00:00<00:00, 62.6MB/s]
Downloading: "https://www.cellpose.org/models/nuclei_3" to /root/.cellpose/models/nuclei_3
100%|██████████| 25.2M/25.2M [00:00<00:00, 44.3MB/s]
Downloading: "https://www.cellpose.org/models/size_nuclei_0.npy" to /root/.cellpose/models/size_nuclei_0.npy
100%|██████████| 3.55k/3.55k [00:00<00:00, 1.55MB/s]
Downloading: "https://www.cellpose.org/models/nucleitorch_0" to /root/.cellpose/models/nucleitorch_0
100%|██████████| 25.3M/25.3M [00:00<00:00, 48.7MB/s]
Downloading: "https://www.cellpose.org/models/nucleitorch_1" to /root/.cellpose/models/nucleitorch_1
100%|██████████| 25.3M/25.3M [00:00<00:00, 62.8MB/s]
Downloading: "https://www.cellpose.org/models/nucleitorch_2" to /root/.cellpose/models/nucleitorch_2
100%|██████████| 25.3M/25.3M [00:00<00:00, 55.3MB/s]
Downloading: "https://www.cellpose.org/models/nucleitorch_3" to /root/.cellpose/models/nucleitorch_3
100%|██████████| 25.3M/25.3M [00:00<00:00, 91.6MB/s]
Downloading: "https://www.cellpose.org/models/size_nucleitorch_0.npy" to /root/.cellpose/models/size_nucleitorch_0.npy
100%|██████████| 3.54k/3.54k [00:00<00:00, 7.07MB/s]
** TORCH CUDA version installed and working. **
>>>> using GPU
import skimage.io
image = skimage.io.imread(file_list2[0])
masks, flows, styles, diams = model.eval([image], diameter=15, channels=[0,0])
processing 1 image(s)
/usr/local/lib/python3.7/dist-packages/torch/nn/functional.py:3829: UserWarning: Default grid_sample and affine_grid behavior has changed to align_corners=False since 1.3.0. Please specify align_corners=True if the old behavior is desired. See the documentation of grid_sample for details.
"Default grid_sample and affine_grid behavior has changed "
time spent: running network 1.58s; flow+mask computation 1.05
estimated masks for 1 image(s) in 2.65 sec
>>>> TOTAL TIME 2.65 sec
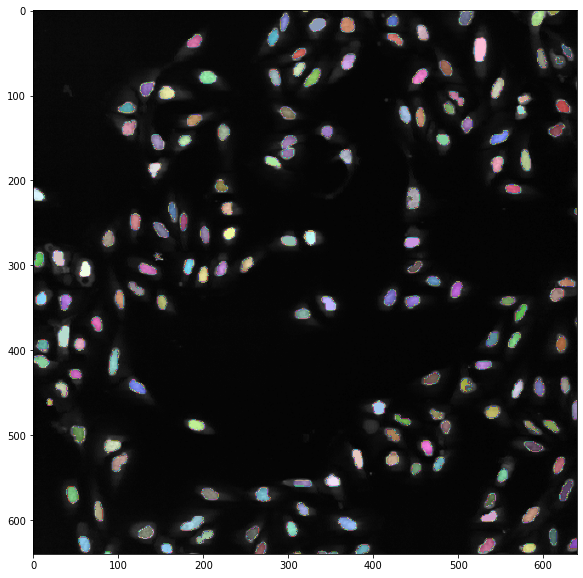
Finally we can look at the output:
from microfilm import microplot
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1,1, figsize=(10,10))
ax.imshow(image, cmap = 'gray', vmax=100)
ax.imshow(masks[0], cmap=microplot.random_cmap(0.5));
Using external software¶
Since we have access to the command line from the notebook, we can both install and run any software from there. Here we show how to do this with Ilastik.
We trained an Ilastik pixel classifier with three images and saved the project. Now we uploaded the project to GitHub to make it available and can use it in this notebook to segment new images.
Installation¶
First we need to install Ilastik. We download it using wget and unzip it:
!wget https://files.ilastik.org/ilastik-1.3.3post3-Linux.tar.bz2
--2021-04-17 19:09:20-- https://files.ilastik.org/ilastik-1.3.3post3-Linux.tar.bz2
Resolving files.ilastik.org (files.ilastik.org)... 194.94.44.60
Connecting to files.ilastik.org (files.ilastik.org)|194.94.44.60|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 370980201 (354M) [application/x-bzip2]
Saving to: ‘ilastik-1.3.3post3-Linux.tar.bz2’
ilastik-1.3.3post3- 100%[===================>] 353.79M 19.3MB/s in 24s
2021-04-17 19:09:45 (14.5 MB/s) - ‘ilastik-1.3.3post3-Linux.tar.bz2’ saved [370980201/370980201]
!tar xjf ilastik-1.3.3post3-Linux.tar.bz2
Now we also download the Ilastik project from GitHub:
!wget https://github.com/guiwitz/ImagingStats/raw/master/ilastik/ilastik_demo.ilp
--2021-04-17 19:13:39-- https://github.com/guiwitz/ImagingStats/raw/master/ilastik/ilastik_demo.ilp
Resolving github.com (github.com)... 140.82.112.3
Connecting to github.com (github.com)|140.82.112.3|:443... connected.
HTTP request sent, awaiting response... 302 Found
Location: https://raw.githubusercontent.com/guiwitz/ImagingStats/master/ilastik/ilastik_demo.ilp [following]
--2021-04-17 19:13:39-- https://raw.githubusercontent.com/guiwitz/ImagingStats/master/ilastik/ilastik_demo.ilp
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 295260 (288K) [application/octet-stream]
Saving to: ‘ilastik_demo.ilp’
ilastik_demo.ilp 100%[===================>] 288.34K --.-KB/s in 0.006s
2021-04-17 19:13:39 (45.2 MB/s) - ‘ilastik_demo.ilp’ saved [295260/295260]
Running ilastik¶
We now need to create a command that will run our analysis. For that we’ll need the location of the ilastik project, the location of the run_ilastik.sh script to run Ilastik and the location one or more images. Then we usse the formatting described in the ilastik documentation to create the command:
import os
from pathlib import Path
ilastik_location = '/content/ilastik-1.3.3post3-Linux/run_ilastik.sh'
ilastik_project = '/content/ilastik_demo.ilp'
output = 'output'
if not os.path.isdir(output):
os.makedirs('output')
command = ('%s --headless --readonly --project="%s" --output_format=tif --export_source="Simple Segmentation"'
' --output_filename_format="%s/{nickname}_results.tif" ') % (
ilastik_location,
ilastik_project,
output)
files_to_process = [x.as_posix() for x in file_list2]
command_full = command+' '.join(files_to_process[0:3])
command_full
'/content/ilastik-1.3.3post3-Linux/run_ilastik.sh --headless --readonly --project="/content/ilastik_demo.ilp" --output_format=tif --export_source="Simple Segmentation" --output_filename_format="output/{nickname}_results.tif" BBBC013_v1_images_bmp/Channel2-25-C-01.BMP BBBC013_v1_images_bmp/Channel2-90-H-06.BMP BBBC013_v1_images_bmp/Channel2-68-F-08.BMP'
import subprocess
subprocess.call(command_full, shell=True)
0
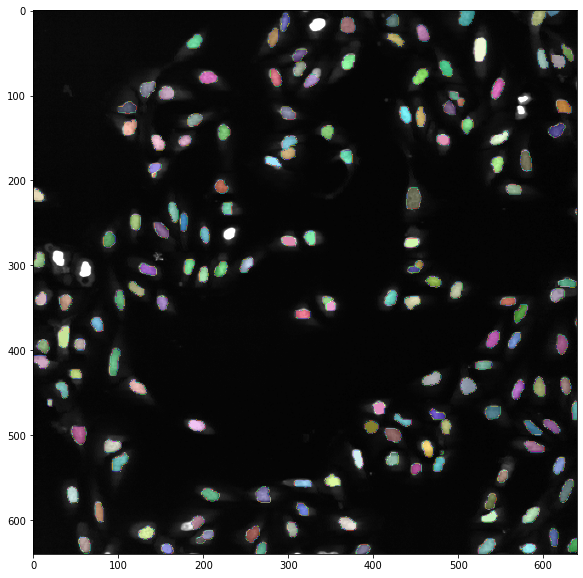
Finally, we can check the result:
import skimage.io
import skimage.morphology
image_to_segment= skimage.io.imread('BBBC013_v1_images_bmp/Channel2-25-C-01.BMP')
segmented = skimage.io.imread('/content/output/Channel2-25-C-01_results.tif', plugin='tifffile')
fig, ax = plt.subplots(1,1, figsize=(10,10))
ax.imshow(image_to_segment, cmap = 'gray', vmax=100)
ax.imshow(skimage.morphology.label(segmented), cmap=microplot.random_cmap(0.5));